| Contributors |
Wiki » History » Version 7
« Previous -
Version 7/8
(diff) -
Next » -
Current version
Adrian Quintana, 21 Mar 2016 11:10
CA1 pyramidal cell - Ferguson et al. 2014¶
Ferguson KA, Huh CY, Amilhon B, Williams S, Skinner FK (2014) Simple, biologically-constrained CA1 pyramidal cell models using an intact, whole hippocampus context. F1000Res 3:104
Izhikevich-based models of CA1 pyramidal cells, with parameters constrained based on a whole hippocampus preparation. Strongly and weakly adapting models based on the experimental data have been developed. Code produces example model output.
NeuroML2/LEMS version of model¶
The model defined in the Python Brian scripts has been converted to a LEMS ComponentType defintion: IzhikevichFerguson.xml.
Note this is different from the existing Izhikevich model definition in NeuroML2, as outlined in Ferguson et al. 2014:
The model has a fast variable representing the membrane potential, V (mV),
and a slow “recovery” current given by the variable u (pA).
In order to capture the spike width at threshold, we slightly modified the
Izhikevich model by using a different “k” parameter above
and below the spike threshold (khigh and klow respectively).
A LEMS file (LEMS_TwoCells.xml) creates a network with 2 instances of the cells:
<izhikevichFergusonCell id="weaklyAdapting" v0 = "-65mV" C="300 pF" vr = "-61.8 mV"
vpeak = "22.6 mV" c = "-65.8 mV" klow = "0.5 nS_per_mV" khigh = "3.3 nS_per_mV"
a = "0.001 per_ms" d = "5 pA" vt = "-57.0 mV" b = "3 nS"/>
<izhikevichFergusonCell id="stronglyAdapting" v0 = "-65mV" C="115 pF" vr = "-61.8 mV"
vpeak = "22.6 mV" c = "-65.8 mV" klow = "0.1 nS_per_mV" khigh = "3.3 nS_per_mV"
a = "0.0012 per_ms" d = "10 pA" vt = "-57.0 mV" b = "3 nS"/>
applies currents as specified in the Python scripts and runs a simulation with the cells:
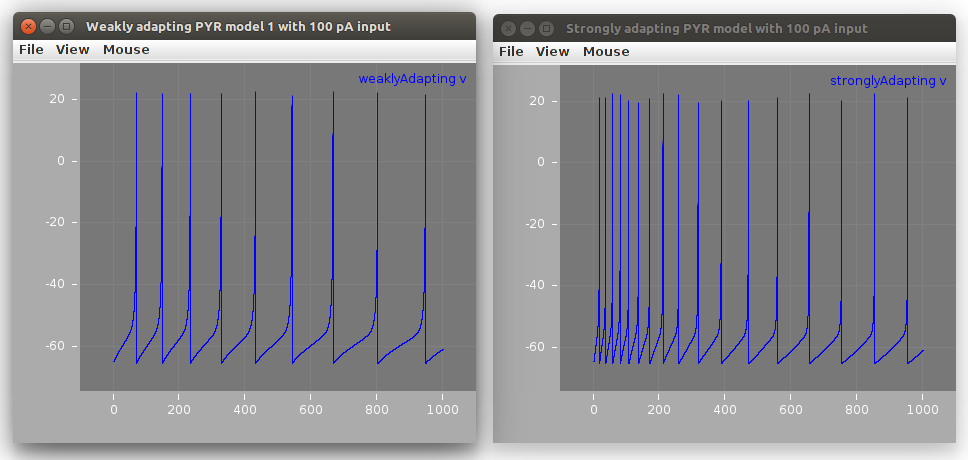
This model can be run locally using jNeuroML:
git clone https://github.com/OpenSourceBrain/FergusonEtAl2014-CA1PyrCell.git # Clone GitHub repository cd FergusonEtAl2014-CA1PyrCell/NeuroML2 jnml LEMS_TwoCells.xml
or using pyNeuroML:
git clone https://github.com/OpenSourceBrain/FergusonEtAl2014-CA1PyrCell.git # Clone GitHub repository cd FergusonEtAl2014-CA1PyrCell/NeuroML2 pynml LEMS_TwoCells.xml
Original Brian scripts for model¶
These files are the original model scripts for use with the Brian simulator as submitted to ModelDB.
To run the scripts, install Brian (version 1) and run:
git clone https://github.com/OpenSourceBrain/FergusonEtAl2014-CA1PyrCell.git # Clone this GitHub repository cd FergusonEtAl2014-CA1PyrCell/Brian python strongly_adapting_PYR_model.py python weakly_adapting_PYR_model.py


