| Contributors |
Cerebellar Granule cell model - Maex and De Schutter, 1998¶
A project containing an implementation of the Granule Cell model from: Maex, R and De Schutter, E.
Synchronization of Golgi and Granule Cell Firing in a Detailed Network Model of the Cerebellar
Granule Cell Layer J Neurophysiol, Nov 1998
For more details see: http://opensourcebrain.org/projects/granulecell
Latest stable release:
Installation¶
To get a copy of this model clone the GitHub repository:
git clone https://github.com/OpenSourceBrain/GranuleCell.git
Run the neuroConstruct project¶
The main content of this project is a neuroConstruct project. More about using neuroConstruct projects here.
Using neuroConstruct, executable scripts for this model can be generated for NEURON, GENESIS and MOOSE.
NeuroML 2 version of model¶
A version of this model in NeuroML2 can be found in the generatedNeuroML2 directory.
This can be run using jNeuroML. Installation instructions here. Use the jnml command line utility to run the model:
cd neuroConstruct/generatedNeuroML2 jnml LEMS_GranuleCell.xml
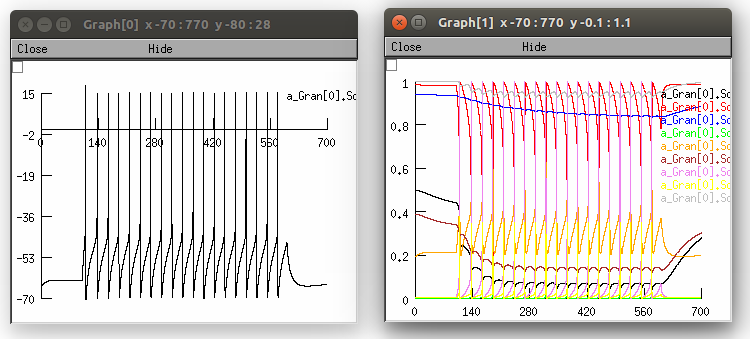
Alternatively, LEMS_GranuleCell_LowDt.xml is a similar model which has a lower timestep for more accurate results in jLEMS, the LEMS simulator embedded in jNeuroML:
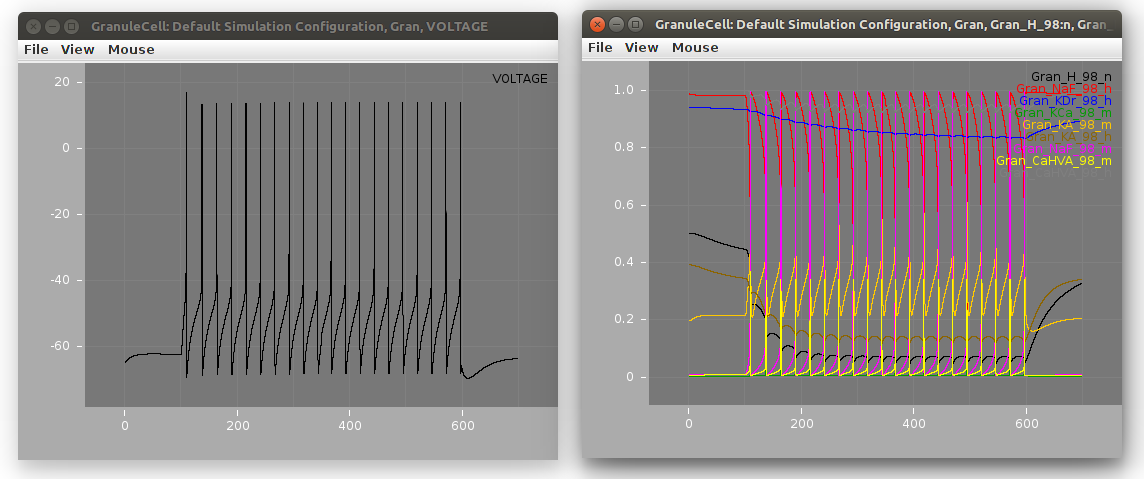
The NeuroML2 version of the model can also be converted to NEURON and run:
jnml LEMS_GranuleCell.xml -neuron nrnivmodl nrngui LEMS_GranuleCell_nrn.py