| Contributors |
|
Example scripts for testing submission to NSG Portal¶
NEURON (hoc format)¶
There is an example in the directory ParallelNetwork which represents a network of 500 cells in NEURON which was generated by neuroConstruct which is set up to run on Parallel NEURON.
You can get a local copy of these files with:
git clone https://github.com/OpenSourceBrain/NSGPortalShowcase.git cd NSGPortalShowcase/examples/NEURON/ParallelNetwork
You should be able to run this locally if NEURON is set up & configured to allow simulations in Parallel:
nrnivmodl mpirun -np 4 nrniv -mpi init.hoc
The exact for of the above command will depend on the version of MPI installed.
To run it on NSG, either use the pre built zip file input.zip, or edit the hoc/mod and rebuild the zip file:
./makezips.sh
and submit this to NSG.
Submitting zip files to NSG¶
- Log in (after having signed up...)
- Create a new folder (e.g. TestFolder)
- Click on Data (0) under the newly created folder on the left
- Click Enter/Upload Data
- Locate input.zip. Name the data TestData.
- Click on Tasks (0); Create New Task
- Name it TestTask.
- Click on Input and select TestData
- Click on Tool and select NEURON7.3 on Comet
- Imput parameters should not need to change. The form should look something like:
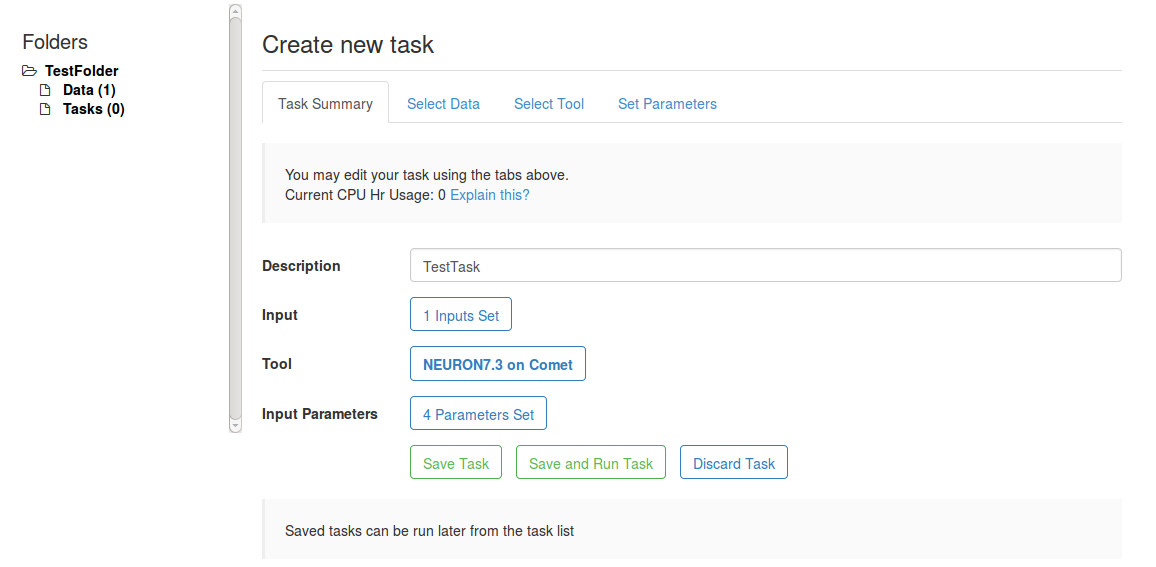
- Hit Save and Run Task.
- Status of ongoing tasks can be seen at Tasks (1) & clicking View Status
- When it finished click View Output and you can download results.
NEURON (Python format)¶
See directory PythonNeuron.
This can be submitted in the same way as above, except use input2.zip and select NEURON7.3 Python on Comet.
Brain¶
See directory Brian.
This can be submitted in the same way as above, except use input.zip and select Brian on Comet.


