Description
point-neuron-fitted-in-Brain¶
About¶
Code to fit single neuron models to Lantyer et al. database (da Silva Lantyer et al., 2018) in Brian . Models are fitted to experimental 'frozen noise' traces (Zeldenrust et al. 2017) using the the Brian simulator (https://briansimulator.org/).
We use the modelfitters available in Brian (https://github.com/brian-team/brian2modelfitting), first the Tracefitter, to fit model voltage traces to the experiments, and then the Spikefitter. Note that for activation particle m of the sodium channel, the half-width Vh and gain k are linearly dependent (see below), so that they do not have to be fitted independently.
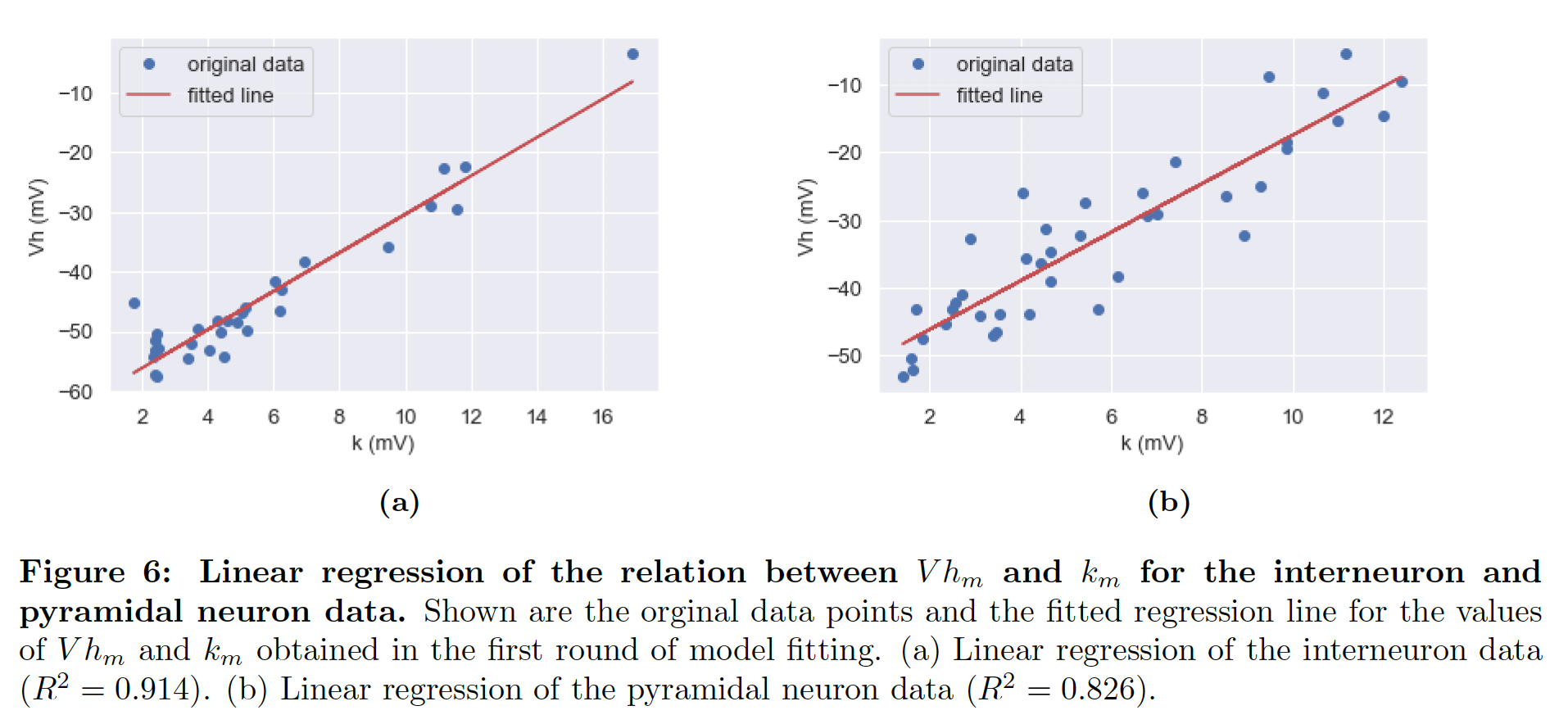
References¶
- da Silva Lantyer, A., Calcini, N., Bijlsma, A., Kole, K., Emmelkamp, M., Peeters, M., Scheenen, W. J. J., Zeldenrust, F., & Celikel, T. (2018). A databank for intracellular electrophysiological mapping of the adult somatosensory cortex. GigaScience, 7(12), 1–9. https://doi.org/10.1093/gigascience/giy147
- Zeldenrust, F., de Knecht, S., Wadman, W. J., Denève, S., & Gutkin, B. S. (2017). Estimating the Information Extracted by a Single Spiking Neuron from a Continuous Input Time Series. Frontiers in Computational Neuroscience, 11, 49. https://doi.org/0.3389/fncom.2017.00049
Members
Scientific Coordinator: Fleur Zeldenrust
References
The code for this model is hosted on GitHub: https://github.com/DepartmentofNeurophysiology/point-neuron-fitted-in-Brian


