Description
Hodgkin Huxley Tutorials¶
This repository contains the code for a number of related tutorials on the Hodgkin Huxley model
Tutorial 1A: HH model in Python and NeuroML¶
Target audience: those who want to learn how to implement the HH model
This tutorial contains a side-by-side comparison of Python code that runs the Hodgkin-Huxley equations and generates plots with a NeuroML2 implementation of the same equations.
This was originally created by @joebowen on behalf of the OpenWorm project.
The latest version of the tutorial is visible online at ReadTheDocs.
Tutorial 1B: Interactive introduction to the HH model in a Jupyter notebook¶
Target audience: those who want to simulate the HH model using an interactive Python based web notebook
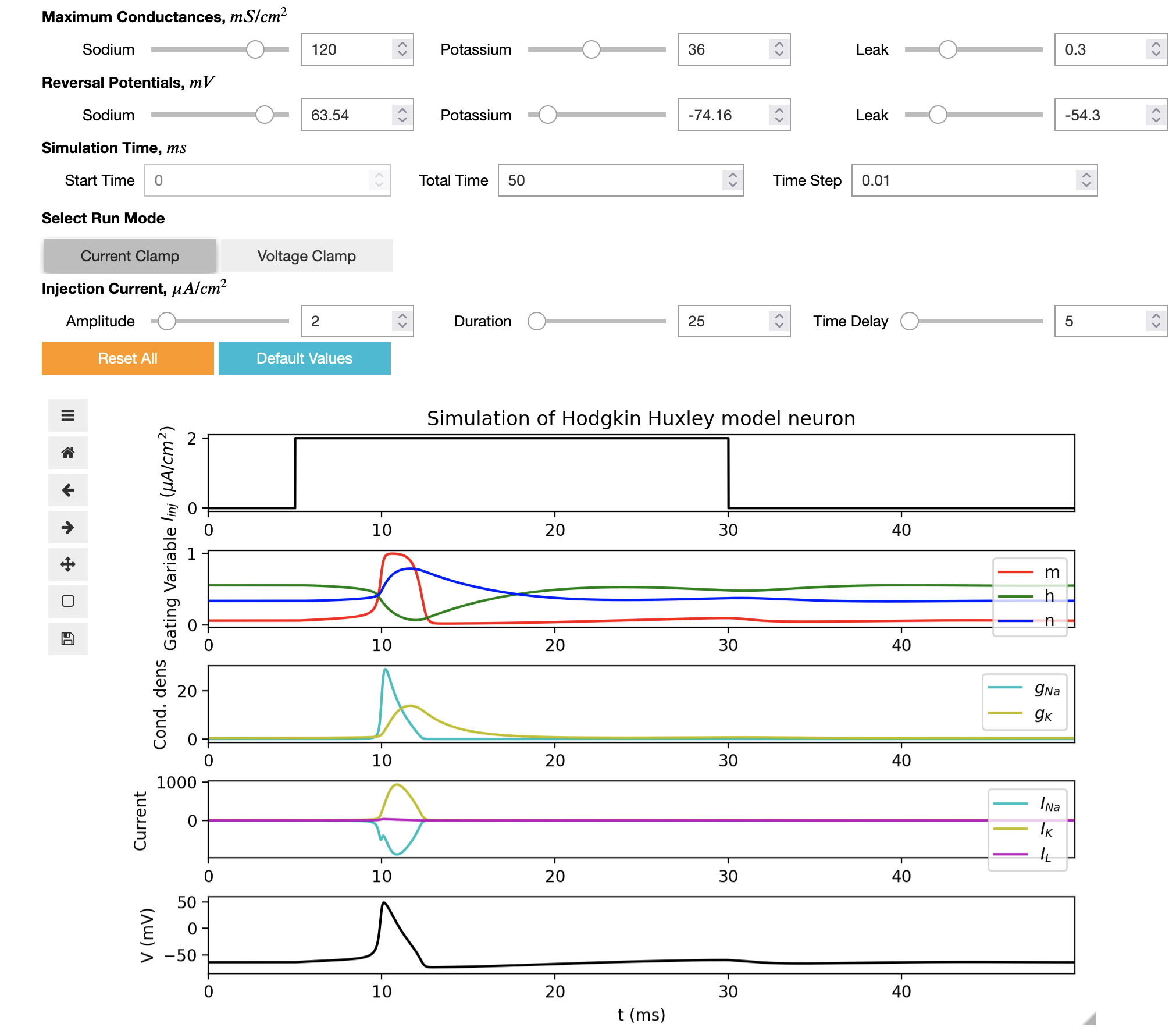
This interactive Jupyter notebook can be used to run the Python version of the HH model from above, change the parameters of the model and display the dynamical properties of variables, without the need to write any code.
Full details can be found here. This work was carried out as part of Google Summer of Code 2022 by Rahul Sonkar.
Tutorial 2: Interactive introduction to the HH model on Open Source Brain¶
Target audience: those who want to learn the basics of the HH model through running in-browser simulations
This tutorial also uses the NeuroML model from above, but provides a high level introduction to the concepts of the model (as well as computational modelling in neuroscience).
It can be accessed on the Open Source Brain site at this location: http://www.opensourcebrain.org/tutorials. It is built on the Geppetto platform, which was also initially developed in the OpenWorm project.
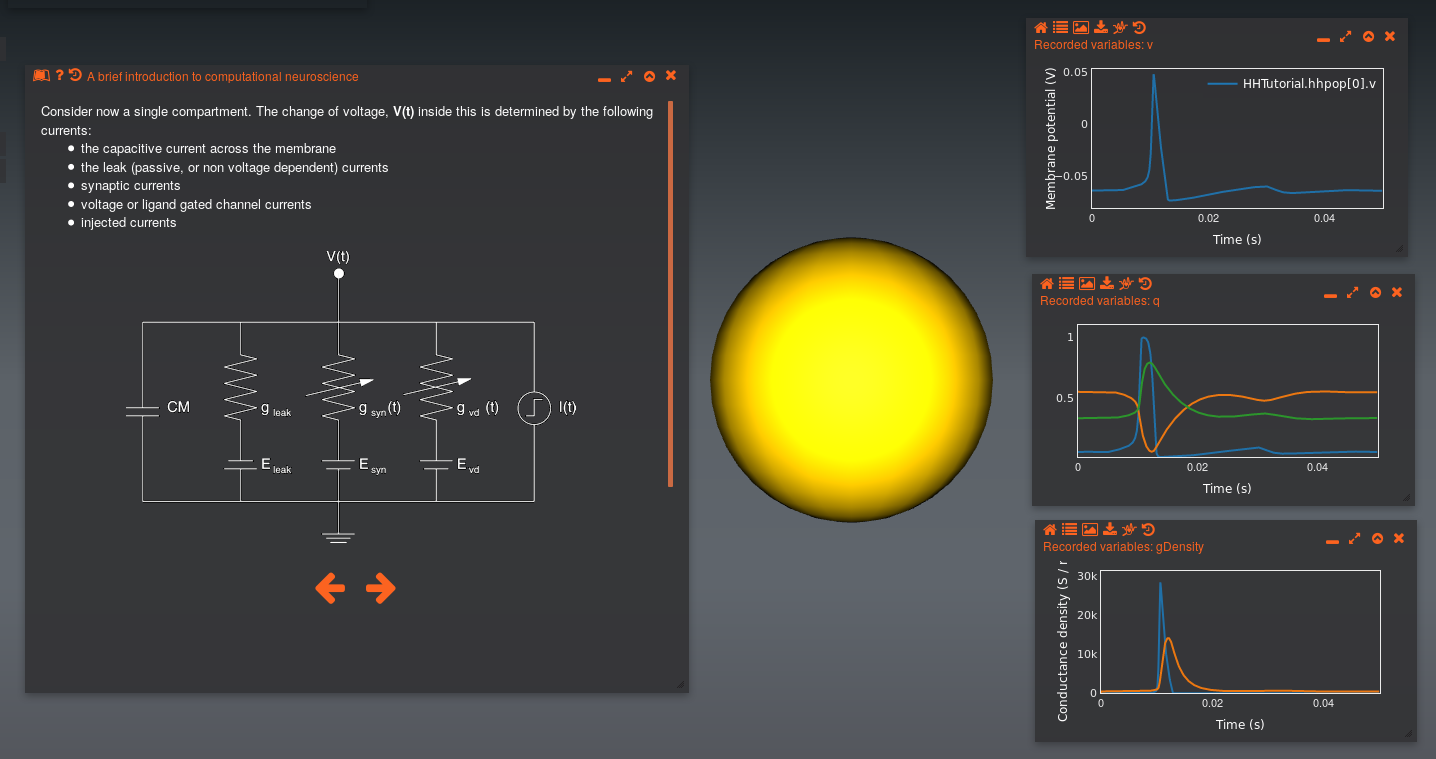
See also http://www.opensourcebrain.org/projects/hodgkin-huxley-tutorial.
Reusing this model¶
The code in this repository is provided under the terms of the software license included with it. If you use this model in your research, we respectfully ask you to cite the references outlined in the CITATION file.
Status
Stable. Developed initially by members of the OpenWorm community.
Members
Developer: Padraig Gleeson, Stephen Larson
Scientific Coordinator: Joe Bowen
References
The code for this model is hosted on GitHub: https://github.com/openworm/hodgkin_huxley_tutorial.git