Description
This is a project implementing the network model of of electrically coupled cerebellar Golgi cells, as described in Vervaeke et al.
Rapid Desynchronization of an Electrically Coupled Interneuron Network
with Sparse Excitatory Synaptic Input, Neuron 2010.
See http://www.opensourcebrain.org/projects/vervaekeetalgolgicellnetwork
for more details.
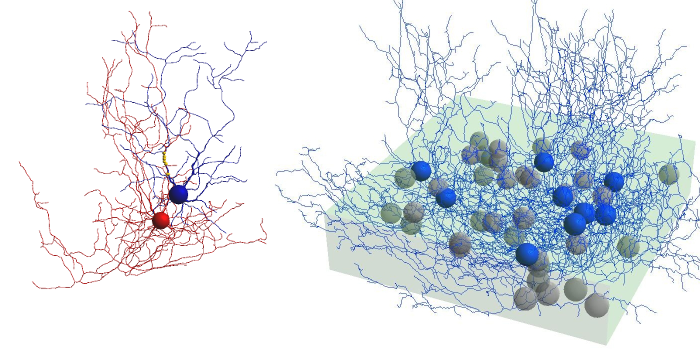
The Golgi cell model is based on the abstract cell model in Solinas et al., Frontiers in Neuroscience 2007.
Reusing this model¶
The code in this repository is provided under the terms of the software license included with it. If you use this model in your research, we respectfully ask you to cite the references outlined in the CITATION file.
Status
Well tested mapping from NeuroML v1.8.1 -> NEURON. KAHP channel with kinetic scheme formalism precludes use on GENESIS/MOOSE for now. Calcium dynamics prevent using this model on PSICS.
Members
Developer: Birgit Kriener, Eugenio Piasini, Miklos Szoboszlay, Padraig Gleeson
Scientific Coordinator: Koen Vervaeke
Scientific Advisor: Angus Silver
References
The original published version of this model is available on

This model was originally developed in: neuroConstruct
The code for this model is hosted on GitHub: https://github.com/OpenSourceBrain/VervaekeEtAl-GolgiCellNetwork


